Genetics
How to Interpret Personal Genetic Testing Reports
The availability of genetic testing is increasing, but is our understanding?
Posted September 11, 2020 Reviewed by Matt Huston

Whether in a medical context, such as prenatal genetic screening, or by sending off a tube of your spit in the mail, information about our own genetics is becoming more easily accessible. However, what does all this information really mean?
The first thing that needs to be clear is that the vast majority of people receiving genetic testing results are not having their whole genome sequenced. Your genome is all the DNA, everything. Like a dictionary, your genome is full of important genes that are like the common words everyone uses all the time. The genome also includes some genes that are rarely if ever important, and even some that seem to have fallen out of usage, such as so-called pseudo-genes, which can be like obscure phrases in old English that nobody would even understand if you used them today. However, your genome also includes a lot of junk, potentially the vast majority of our DNA, with no particular purpose.
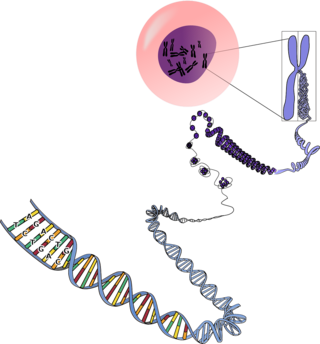
It is possible that the regions outside of genes and known regulatory sequences have some function we don’t yet understand, but for now, there is a general consensus that if you want to find out about genetic diseases, it might be sufficient to look first at all the DNA in the genome that actually makes up genes. This is referred to as the exome because it is all the sequences that are “expressed.” However, most genetic testing isn’t even based upon whole-exome sequencing.
Broadly speaking, when we look at the results of a genetic test, the information provided comes in one of two different formats. In one paradigm, individual genes are analyzed, or even small portions of single genes known to harbor potentially impactful mutations. Alternatively, genetic tests can include information about the identities of thousands of small variants, none of which has a particularly strong functional impact.
These two alternative approaches stem directly from the fact that although some so-called monogenic genetic diseases arise from single mutations with very significant functional impacts, the majority of diseases with a genetic component do not. Rather, the combined impact of many subtle genetic variants, placed within specific environment conditions, result in an increased susceptibility relative to the general population. These variants are collectively referred to as single nucleotide polymorphisms (SNPs), and the patterns of SNPs you inherit correspond with the risk of developing this type of complex so-called polygenic genetic disease. The application of SNP data to understanding an individual's potential for developing a particular disease with a genetic component can be calculated into what is referred to as a polygenic risk score—however, this is rarely a sure thing and generally depends upon other genetic and environmental factors.
Alternatively, if you inherit two copies of the so-called delta-F508 mutant variant of the gene CFTR, you will develop the monogenic lung disease cystic fibrosis. So prenatal genetic tests routinely look to see if an individual is a carrier of this specific mutation, among many others. However, individual SNPs don’t have this kind of direct functional significance and they are only really useful when looked at in large numbers—like thousands at once.
In many ways, the use of SNPs to understand genetic disease risk is more about statistical associations than direct causality. The technique known as Genome-Wide Association Study (GWAS) underlies much of what informs personal genetic testing. Basically, GWAS looks at thousands of SNPs from people with and without a specific disease that has a genetic component and permits the discovery of specific SNP patterns that correspond to increased risk of developing the disease. GWAS can also be performed within a single study population without a negative control group, routinely including hundreds of thousands of people, looking at a so-called continuous variable that falls into different quantifiable levels, like height, degree of anxiety, or educational attainment.
SNP patterns associated with these differences can then be used for screening purposes. However, this must always be taken into the context of the specific characteristics of the population analyzed as well as the impact of environmental variables. As described by recent work published by Crouch and Bodmer, understanding the actual functional significance of particular SNPs found within GWAS, beyond a calculated polygenic risk score, is not simple. How then can we make well-informed medical or behavioral decisions?
If you find out from a personal genetic test that your specific SNP patterns indicate higher than average risk for heart disease, you might decide to eat a healthier diet and exercise more. However, if you find out that you are not in a high-risk group and you decide to take up smoking and eat bacon cheeseburgers three times a day, you will likely still suffer health impacts from those behavioral choices.
These risk calculations can change over time though, and in the context of single-gene analyses, what is referred to as a variant of uncertain significance today might turn out to be strongly associated with increased risk, or possibly completely innocuous. This is all a work in progress, and unfortunately, while many genetic tests performed in a clinical context are not reanalyzed as further information becomes available, some personal genetic testing companies helpfully do update client reports over time as new discoveries are made.
One major issue with some of these analyses, though, is that historically, many of the populations being studied are relatively genetically homogeneous, that is to say, people of European/white descent. Context can matter a great deal, and variants shown to be indicative of significant risk in one population might not be the same within a different genetic background. Thus, as with many things, increasing diversity within these study populations will vastly improve the usefulness of the outcomes.
So these types of genetic analyses can be extremely beneficial, for example, in identifying carriers of debilitating diseases in prenatal screening, but they are not a panacea. Increased susceptibility doesn’t mean that something will definitely occur. Furthermore, even in the absence of a particular risk factor, you still may develop a particular disease, especially if it depends on many different genetic and behavioral variables. Finally, our understanding is always changing. The analysis of more people, particularly from diverse genetic backgrounds, is required, as is a better understanding of the biological and environmental causes of disease. Thus, for now, these tests can be extremely valuable, as long as we know what questions are being asked, and what the answers truly mean and don’t mean.




